A New Tool in the Toolbox: New Method Traces Free-Floating DNA Back to Its Source
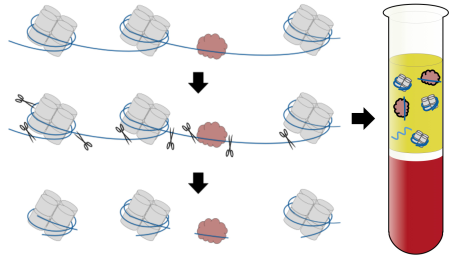
Caption: DNA (blue) loops around nucleosomes (gray) and is bound by transcription factors (red), proteins that switch genes on and off and act in a tissue-specific manner. When cells die, enzymes (scissors) chop up areas between the nucleosomes and transcription factors, releasing DNA fragments in unique patterns. By gathering the released DNA fragments in blood, researchers can tell which types of cells produced them.
Credit: Shendure Lab/University of Washington
Credit: Shendure Lab/University of Washington
When cells die, scissor-like enzymes snip their DNA into tiny fragments that leak into the bloodstream and other bodily fluids. Researchers have been busy in recent years working on ways to collect these free-floating bits of DNA and explore their potential use in clinical care.
These approaches, sometimes referred to as “liquid biopsies,” hinge on the ability to distinguish specific DNA fragments from the body’s normal background of “cell-free” DNA, most of which comes from dying white blood cells. Seeking other sources for cell-free DNA in particular situations is beginning to bear fruit, however. Current applications include: 1) a test in maternal blood to look for DNA from the fetus (actually from the fetal component of the placenta), which provides a means of detecting a possible genetic abnormality; 2) a test in a cancer patient’s blood to look for cancer-specific mutations, as a way of assessing response to treatment or early signs of relapse; and 3) a test in an organ transplant recipient, where increasing abundance of DNA fragments from the donor can be an early sign of rejection.
But recent proposals have been floated about looking for cell-free DNA in healthy individuals, as an early sign of some health problems. Suppose something was found—how could you know the source? Now a team of NIH-funded researchers has devised a new method that uses distinctive features of DNA packaging to provide an additional layer of information about the origins of free-floating DNA, vastly expanding the potential uses for such tests [1].
The new cell-free DNA method, described in a recent proof-of-concept study in the journal Cell, opens the possibility of one day identifying the likely source of invasive “cancers of unknown primary” with a simple blood draw. The method might also improve the diagnosis, treatment, and management of a vast array of health conditions that lead to an abnormal increase in cell death and cell-free DNA, including heart attack, stroke, and autoimmune diseases such as rheumatoid arthritis and lupus.
Jay Shendure, along with graduate student Matthew Snyder and postdoctoral fellow Martin Kircher, at the University of Washington, Seattle, developed the new approach for analyzing cell-free DNA. Their method capitalizes on the fact that DNA within cells is wound around protein complexes to form millions of compact structures called nucleosomes, often likened to beads on a string.
The precise locations of nucleosomes (the beads) and the lengths of the spaces between them (the string), differ among cell types, because of the presence of cell-specific proteins called transcription factors that regulate gene expression and interrupt the monotonous placement of nucleosomes along the DNA strand. In fact, this packaging of DNA into nucleosomes helps to explain how the various cells in our bodies can look and behave differently despite being genetically identical. The packaging also suggests that different types of cells hide different—and perhaps distinctive—segments of DNA in nucleosomes.
Those are important points, because the DNA-degrading enzymes are less likely to make cuts in the DNA wound tightly around the nucleosomes. They will cut instead in between them. And when a transcription factor is present, that pattern will be reflected by the cut sites. For that reason, Shendure’s team suspected that most of the cell-free DNA fragments in blood should come from nucleosomes and bear distinctive segments of DNA from the cells that produced them. If they were correct, the team members could match cell-free DNA sequences to existing maps showing the locations of nucleosomes in various cell types, allowing them to trace the loose DNA fragments to their most likely cellular sources.
To test their hypothesis, the researchers first sequenced more than 3 billion fragments of cell-free DNA isolated from healthy people. They were able to determine that the distribution of fragments was indeed consistent with cleavage along the edges of nucleosomes. They also found in 116 blood samples that the predicted spacing of nucleosomes was consistent with cell-free DNA derived from white blood cells, as would be expected in healthy people under normal circumstances. They could even tell where transcription factors had been that regulate gene expression in cells normally present in the blood.
To take their proof-of-concept a step further, the researchers examined blood samples from five patients with various forms of advanced cancer. In contrast to what they’d seen in healthy people, these patients had cell-free DNA that often traced back to the organ that gave rise to their cancer. For example, the cell-free DNA from a person with a common form of liver cancer suggested a pattern of nucleosome spacing most consistent with a liver cancer cell line. The predicted pattern of nucleosomes in cell-free DNA from a person with breast cancer most closely resembled that of a metastatic breast cancer cell line.
The new findings show that it’s possible to glean another layer of useful information from cell-free DNA tests. While the new method will need to be evaluated in many more samples and under many more physiological conditions, it raises the possibility that cell-free DNA tests could be applied to a much broader range of conditions, especially those marked by acute or chronic tissue damage. The researchers say they are continuing to explore the method’s possible applications. They’ll also be exploring ways to convert their analytical framework into an affordable test.
Reference:
[1] Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell. 2016 Jan 14;164(1-2):57-68.
Links:
Cancers of Unknown Primary (National Cancer Institute/NIH)
Jay Shendure Lab (University of Washington, Seattle)
NIH Support: Common Fund


































No hay comentarios:
Publicar un comentario