
NCI Expands Repository of Cancer Research Models: A Conversation with Drs. Doroshow and Evrard
April 20, 2018, by NCI Staff
NCI’s Patient-Derived Models Repository (PDMR) is an initiative to generate and distribute cancer research models made directly from patients’ tumor tissue for use in laboratory research. NCI recently expanded the PDMR to include more research models, including models of some rare cancers.
In this interview, Yvonne A. Evrard, Ph.D., PDMR program lead at the Frederick National Laboratory for Cancer Research, and James H. Doroshow, M.D., director of NCI’s Division of Cancer Treatment and Diagnosis, discuss why these models are needed and the unique characteristics of the repository.
What was the impetus behind the development of the PDMR?
Dr. Evrard: In 2012, Dr. Doroshow pulled together a meeting with leaders in cancer research from academia and pharmaceutical companies to figure out what was needed to advance preclinical cancer research.
Meeting participants said there was a major need for patient-derived models—laboratory models that more closely resemble human tumors than traditional cancer cell lines, which have been grown in plastic dishes for many years. A lot of genetic changes can occur when you grow cells in the laboratory for an extended period. While traditional cell lines have huge value in preclinical research, we need models that are one step closer to the patient—models that are more predictive of treatment outcomes.
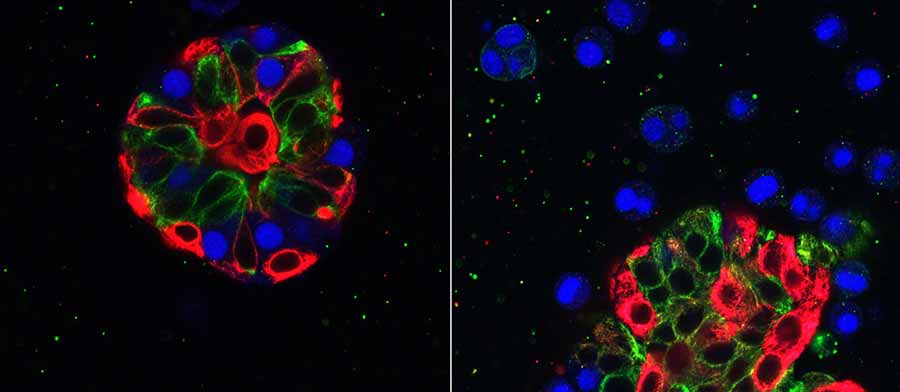
The PDMR currently includes patient-derived xenograft (PDX) models, which are made by taking a sample of a patient’s tumor and grafting it into a mouse. Many studies have shown that these models are better able to mimic the patient’s therapeutic outcomes.
Dr. Doroshow: The primary goal of this repository is to make a publicly available archive and repository of tumor samples and models that are fully characterized from a molecular and clinical perspective, and that are easily accessible to the extramural community at a very modest cost.
All of the models in the PDMR are clinically annotated. What does that mean, and why is it important to cancer researchers?
Dr. Evrard: Clinically annotated means we collected relevant information about the patient whose tumor sample was used to create the model. That includes their cancer treatment and outcome history, self-reported race and ethnicity, smoking history, and occupation. Knowing that a model is developed from a patient who has not received treatment compared with a patient who has, for example, helps researchers select models that might have sensitivity or resistance to a specific drug, or to look for biomarkers linked to those properties.
In addition to collecting information on self-reported race and ethnicity, we do genetic ancestry testing on all tumor samples. There are a lot of cases where specific cancers have different outcomes in patients of different racial and ethnic backgrounds, and in some cases, this has been tied to the genetic components of their ancestry. We also collect patient occupation history because certain occupations have more exposure to harmful chemicals than others.
What new models have been added to the repository?
Dr. Evrard: We recently released models of several tumor types, including models of some rare cancers. We added gynecologic cancers, osteosarcomas, and small cell lung cancers, among others. The clinical data are currently available for download, and the models are available on request.
How is the NCI PDMR unique from other cancer model collections?
Dr. Doroshow: One key difference is that the PDMR will eventually have different types of models. We are working to provide solid tumor samples for PDX models, and, whenever possible, to develop conditionally reprogrammed cell (CRC) lines and organoid models from patient tissues.
CRCs are patient tumor cells that have been modified so that they can be grown in laboratory dishes. In the past, it has been difficult to grow patients’ cells in cell culture. But, over the past few years, we’ve learned how to successfully establish cancer cell lines from patient tumor samples using this conditional reprogramming approach, and these cell lines are proving to be very valuable research tools, including for testing potential new cancer drugs.
Organoids are another kind of laboratory model. They are three-dimensional structures that mimic the organs or tissues from which they originated—in this case, tumors.
Both of these types of models better represent the structure and complexity of human tumors than traditional cell lines do. The goal is to provide cell cultures for researchers to do initial biological or drug screening—which is less expensive than testing in mice—and if they get interesting results, they could then confirm the findings in PDX mouse models of the same tumors.
We also have close to 100 cell lines made from tumor-associated fibroblasts—connective tissue cells that are an important component of the tumor microenvironment. They will soon be added to the PDMR so the research community can study the interactions between tumor cells and cancer-associated fibroblasts. In some cases, we have a tumor cell line and fibroblast cell line from the same patient, so investigators will be able to study the interactions between tumor cells and fibroblasts from a specific patient.
Having these tumor-associated fibroblast cell lines will enable researchers to get a better picture of how tumor cells behave in actual tumors.
When the tumor tissue is available, we are also developing many of these model types from both the primary tumor and multiple metastatic tumors from the same patient, so researchers can study the process of metastasis.
Dr. Evrard: Another important difference between the PDMR and other repositories of cancer models is that we do genetic sequencing and RNA sequencing of all our models. But we don’t just do them on a single PDX, we’re sequencing multiple tumors from each PDX model and making all of the data public. The goal is to give researchers a picture of the heterogeneity that exists within those tumors.
An analysis of PDX models in NCI’s repository showed that the models retain key mutations that are found in the patient tumors from which they were derived. So we know that the models closely resemble the original tumors.
Your goal with the PDMR is to generate models with biological and clinical diversity. What does that mean?
Dr. Evrard: Some cancer types, such as breast cancer, have several subtypes of the disease. And between subtypes there is a lot of variation in the genetic makeup and how the tissue appears under a microscope, or the histology.
Diversity also applies to the patients who contribute tissue to PDMR. Preclinical models from racial and ethnic minorities are underrepresented in other collections, so we’re trying to build that diversity into this collection as much as possible.
For example, NCI funded six minority-underserved NCI Community Oncology Research Program sites to collect tissue to be used to generate PDMR models. These sites were specifically selected because of the diversity of their patient populations. We also are communicating to researchers in general that this is something we are looking to expand and prioritize, in the hopes that they’ll submit models developed from the tissue of minority patients.
The other gap we currently have is pediatric cancer models. So we are collaborating with NCI’s Pediatric Oncology Branch to gather the tissue samples needed to develop more models of childhood cancers. Pediatric and adult cancers are very different at the molecular level and they have different responses to therapy. Having better models will allow researchers to better understand these differences and, hopefully, develop better and safer therapies.
Our goal is to generate 1,000 PDX models. We recognize that even 1,000 models are not enough to capture all of this diversity, but it’s a good starting point.
What are some of the intended uses for these models?
Dr. Evrard: In September 2017, NCI launched the PDX Development and Trial Centers Research Network (PDXNet), a consortium of four centers and a data coordinating center, to try to scale up preclinical drug testing in PDX models.
Each of the centers have developed large numbers of PDX models. Through this network, we can take a drug or drug combination and test it in PDX models of multiple cancer types.
Having large collections of models will allow researchers in the network to pick specific groups of tumors for experiments. For example, maybe they want to test a drug in tumors with a specific genetic mutation and they don’t care what the cancer type is. We’ll be making the data from these experiments available so researchers know which models respond to which drugs.
Dr. Doroshow: The PDXNet consortium is going to play a critical role in helping us develop combinations of investigational agents that will then be studied in NCI’s phase 1 clinical trials program. We hope to have a seamless process of identifying drug combinations that we want to test in these trials.The goals are to get agents from our pharmaceutical partners, to study those agents in the PDMR models, and then to develop clinical trials directly from the results that we observe. I don’t believe any other noncommercial venture of this scope exists. And these results are all going to be made public.
Dr. Evrard: And once we have large numbers of models, we can also analyze the genetics to see if we can find unique or new DNA or protein biomarkers —similar to how researchers have analyzed genomic data from The Cancer Genome Atlas. For those types of data mining studies you need a large number of genetic sequences from a large number of tumors. We do have investigators who are using data from the PDMR in that way—they download the genetic sequencing and histology data for bioinformatic analyses, but don’t use the models in a laboratory setting.
Has the research community been using the PDMR?
Dr. Evrard: We have been distributing models to researchers since May of 2017, and we have had groups come back for additional models. There’s huge interest in the CRC and organoid models that will soon be coming out. I’ve heard a lot of positive feedback from the community.
Dr. Doroshow: We have more than 150 PDX models as of April 2018. We’ve sent out many models to academic investigators. We’ve also sent models to pharmaceutical and biotech companies. And we have investigators who are just downloading the molecular and clinical characteristics for data-mining research.



































No hay comentarios:
Publicar un comentario