
Volume 24, Number 6—June 2018
Research
Novel Parvovirus Related to Primate Bufaviruses in Dogs
On This Page
Vito Martella , Gianvito Lanave, Eszter Mihalov-Kovács, Szilvia Marton, Renáta Varga-Kugler, Eszter Kaszab, Barbara Di Martino, Michele Camero, Nicola Decaro, Canio Buonavoglia, and Krisztián Bányai
, Gianvito Lanave, Eszter Mihalov-Kovács, Szilvia Marton, Renáta Varga-Kugler, Eszter Kaszab, Barbara Di Martino, Michele Camero, Nicola Decaro, Canio Buonavoglia, and Krisztián Bányai
Abstract
A novel protoparvovirus species, related genetically to human bufaviruses, was identified in dogs with respiratory signs. The canine bufavirus was distantly related to the well-known canine protoparvovirus, canine parvovirus type 2, sharing low amino acid identities in the nonstructural protein 1 (40.6%) and in the capsid protein 1 (33.4%). By screening collections of fecal, nasal, and oropharyngeal samples obtained from juvenile dogs (<1 year of age), canine bufavirus DNA appeared as a common component of canine virome. The virus was common in the stool samples of dogs with or without enteric disease and in the nasal and oropharyngeal swab samples of dogs with respiratory signs. However, the virus was not detected in nasal and oropharyngeal swab samples from animals without clinical signs.
Parvoviruses (family Parvoviridae) are small, nonenveloped viruses of 25–30 nm in diameter, with an icosahedral capsid. The genome is a single-strand DNA of 4.5–5.5 kb (1) with complex hairpin structures at the 5′ and 3′ ends. The genome is predicted to encode 3 or 4 proteins: nonstructural (NS) 1, nucleoprotein (NP) 1, and viral protein (VP) 1 and 2.
Parvoviruses have long been known in dogs, since the identification of canine minute virus, or canine parvovirus (CPV) type 1 (CPV-1; genus Bocaparvovirus), in 1967 from the fecal samples of healthy dogs (2). CPV-1 infection is responsible for reproductive disorders and occasionally for respiratory and gastrointestinal signs in young puppies (3). A second CPV (CPV-2; genus Protoparvovirus) was reported in the 1970s in Europe and North America in puppies with signs of hemorrhagic gastroenteritis and myocarditis (4). CPV-2 is currently regarded as the major causative agent of severe gastroenteritis in puppies and is included in canine core vaccination schedules globally (5). In 2011, a second canine bocaparvovirus (CBoV) was identified from nasal swabs of healthy and sick dogs (6); a third species of CBoV was identified in 2013 in the liver of a dog with multiorgan failure (7) (Table). Whether the newly identified parvoviruses play a role as canine pathogens has not yet been assessed.
We report the identification of a novel CPV. We determined the genome sequence of the CPV and designed specific primers and probes useful for laboratory diagnosis. Screening of enteric and respiratory samples from dogs with either gastroenteric or respiratory disease and from animals without clinical signs suggested a possible association between the novel virus and respiratory disease in young dogs.
Identification of the DNA of a Novel Parvovirus
In 2011, an outbreak of canine infectious respiratory disease (CIRD) occurred in a litter of 3 mixed-breed 5-month-old puppies in Italy. The animals’ clinical signs were nasal discharge, coughing, and respiratory distress, but they completely recovered from the disease after 2 weeks. Nasal and oropharyngeal swab specimens tested negative to a panel of molecular assays for CIRD-associated common and emerging viral agents: canine adenovirus (CAV) types 1 and 2, canine distemper virus, canid herpesvirus 1 (CHV-1), canine respiratory coronavirus, influenza virus, canine parainfluenza virus, canine pneumovirus, nonprimate canine hepacivirus, B. bronchiseptica, Streptococcus equi subspecies zooepidemicus, and Mycoplasma cynos (12). Because the etiology of the outbreak was unknown, the case was considered eligible for metagenomic investigation.
We performed random primed reverse transcription PCR and PCR assays on pooled samples (the nasal and oropharyngeal swab specimens of the 3 puppies) to amplify nucleic acids and used them as templates for next-generation sequencing (NGS) experiments on the Ion Torrent platform (New England Biolabs, Ipswich, MA, USA) (13). We evaluated sequence data using CLC Genomic Workbench (http://www.clcbio.com). NGS revealed the presence of parvovirus-related sequence reads that mapped to human bufaviruses. We mapped a total of 3,530 reads to human bufavirus and assembled them into 3 contigs, 422-, 416-, and 191-nt long. We reconstructed the nearly complete genomic sequence of the new CPV (canine bufavirus [CBuV]), strain ITA/2011/297-15, by combining 5′ rapid amplification of cDNA ends (RACE) protocols (14) with minor modifications, using the kit 5′ RACE System for Rapid Amplification of cDNA Ends version 2.0 (Life Technologies, Paisley, UK) and a primer-walking strategy with specific primers designed to close the gaps among noncontiguous sequences. We purified and cloned the amplicons using a TOPO XL Cloning Kit (Life Technologies) and generated consensus sequences by sequencing >3 clones for each PCR fragment.
Screening of Samples in Conventional and Quantitative PCR
We designed specific primers on the VP2 genomic region of strain ITA/2011/297-15 for quantitative detection in real-time PCR (qPCR) (CPPV-L3-for 5′ TGAACAAGAAATAGACAACATTGTCAT 3′, CPPV-L3-rev 5′ AAAGAGCAGTTAGGTCATTGTTGT 3′, and CPPV-L3 Pb 5′ Fam CCAAACAAGGTACAGGACAGGAAGAAACAACACAA BHQ1 3′). We calculated CBuV DNA copy numbers on the basis of standard curves generated by 10-fold dilutions of a plasmid standard TOPO XL PCR containing a 500-nt fragment of the VP2 region of strain ITA/2011/297-15. We added 10 μL of sample DNA or plasmid standard to the 15-μL reaction master mix (IQ Supermix; Bio-Rad Laboratories SRL, Segrate, Italy) containing 0.6 μmol/L of each primer and 0.2 μmol/L of probe. Thermal cycling consisted of activation of iTaq DNA polymerase at 95°C for 3 min, 42 cycles of denaturation at 95°C for 10 s, and annealing extension at 60°C for 30 s. We evaluated the specificity of the assay with a panel of canine DNA viruses (CPV-1, CPV-2, CHV-1, circovirus, CAV-1, and CAV-2). The qPCR detected >101 DNA copies/10 μL of standard DNA and 8.78 × 100 DNA copies/10 μL of DNA template extracted from clinical samples. CBuV quantification had an acceptable level of repeatability over various magnitudes of target DNA concentrations, when calculating (15) the intraassay and interassay coefficients of variation within and between runs, respectively.
In addition, we designed specific primers (CPPV 165F 5′ CTGGTTTAATCCAGCAGACT 3′ and CPPV 371R 5′ TGAAGACCAAGGTAGTAGGT 3′) to amplify and sequence a 207-nt fragment of VP2. We used the AccuPrime Taq DNA polymerase (Life Technologies) for PCR amplification. Cycling thermal conditions included initial activation of the polymerase at 94°C for 2 min, 45 cycles at 94°C for 30 s, 53°C for 30 s, and 72°C for 30 s, followed by final extension at 72°C for 10 min.
Respiratory Samples
During 2011–2015, we obtained nasal and oropharyngeal swab (NOP) samples from 58 pups and young dogs (<1 year of age) with CIRD (collection RIS); the animals were early acute clinically ill CIRD dogs with onset of respiratory signs at 0–3 days at the time of sample collection. The RIS samples were a subset of a larger collection used for surveillance of traditional and emerging pathogens associated with CIRD (12). We collected samples from the nasal and oral cavities in parallel and stored them separately. We also screened NOP swabs obtained from 90 dogs <1 year of age without clinical signs (collection RIA) as controls. We collected all samples with dry swabs and immediately stored them at −20°C.
Enteric Samples
We screened archived enteric samples (stools and rectal swabs) collected at the Department of Veterinary Medicine, University of Bari, Italy, during 2010–2015 for CBuV. The samples had been obtained from pups and young dogs (<1 year of age). We screened 81 samples from animals with signs of gastroenteritis (collection EIS) and 78 samples of animals without clinical signs (collection EIA).
We also searched for CBuV DNA in a collection of enteric samples from pups and young dogs (<1 year of age) either with gastroenteritis (collection EHS) or without clinical signs of gastroenteritis (collection EHA), obtained in Hungary in 2012 and available at the Institute of Veterinary Medical Research, Hungarian Academy of Science, Budapest. We screened 40 samples from healthy animals and 20 samples from animals with clinical signs.
Statistical Analysis
We evaluated the associations among clinical signs, age, and presence of the virus in the respiratory and enteric samples by the χ2 test using a web-based software program (R version 3.3.0; http://www.r-project.org/). We set statistical significance to p<0.05.
Genome Analysis of Hungarian CBuV Strains
We selected samples positive for CBuV on the basis of the genome copies as revealed by quantitative PCR. We also identified the genomes of 2 additional CBuV strains, HUN/2012/22 and HUN/2012/126.
Sequence and Phylogenetic Analyses
We retrieved genome sequences of protoparvovirus strains from GenBank and aligned them using the Clustal Omega tool from the European Molecular Biology Laboratory (https://www.ebi.ac.uk/Tools/msa/clustalo/). We conducted sequence and phylogenetic analyses in Geneious version 9.1.8 (Biomatters Ltd., Auckland, New Zealand); we used the neighbor-joining method, Jukes–Cantor genetic distance model, and bootstrapping for <1,000 replicates.
Virus Cultivation
We homogenized PCR-positive respiratory and fecal samples in 10% Dulbecco’s modified Eagle’s medium and then centrifuged them at 10,000 × g. We filtered the supernatant with 0.22-μm filters and inoculated it onto freshly seeded canine fibroblastic tumor (A-72) cells, incubated at 37°C in 5% CO2. We also inoculated Madin-Darby canine kidney cell (MDCK) and Walter Reed canine cell (WRCC) lines. We evaluated viral growth through 5 serial passages, monitoring the onset of cellular cytopathic effect and testing the cell supernatant by qPCR.
Genome Analysis of Canine Bufaviruses
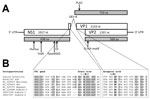
Figure 1. Genome organization of canine bufavirus. A) Positions of the conserved helicase Walker A (GxxxxGKS), Walker B (EE), and replication initiator motifs (HuHuu and YuxK) in NS1 and of the phospholipase A2...
We determined the nearly complete genome (4,537 nt) of the CBuV strain ITA/2011/297-15, including a partial 5′ untranslated region (UTR) (310 nt), the complete NS1 sequence (638 aa), the complete VP1 (710 aa) and VP2 (568 aa) sequences, and a partial 3′ UTR (8 nt). The genome coding sequence, excluding the terminal UTR regions, was 4,219 nt (GenBank accession no. MF198244). The genome contained 2 major open reading frames (ORFs); the left ORF, coding for NS1, was 1,917 nt and the right ORF, encoding VP1 and VP2, was 2,316 nt (Figure 1, panel A). Full-genome sequence alignment showed a high degree of sequence divergence, up to 58% overall nucleotide identity to most parvoviruses but not bufaviruses. CBuV was more closely related to bufaviruses identified in primates (61.6%–63.2% nt similarity), pigs (59.6% nt), and bats (58% nt) (16–20) and more distantly related to CPV-2 (45% nt) (Technical Appendix[PDF - 138 KB - 1 page] Table). The putative bufavirus NS1 start codon was located in a strong Kozak sequence, ACCATGG. The ATP- or GTP- binding Walker loop motif (GXXXXGK[T/S]) was found in NS1 (405-GPASTGKS-412) (21). In addition, the NS1 contained 2 conserved replication initiator motifs, GLHFHVLLQ and IVRYFLTKQP (boldface type indicates conserved amino acids) (22). We generated an nt and aa sequence identity matrix. NS1 showed <69.4% nt and 51.4% aa identity with other parvovirus NS1 sequences, including its closest relatives in the Parvovirus genus (Technical Appendix[PDF - 138 KB - 1 page] Table).
The termination of ORF1 overlapped the start of ORF2 by 14 nt. From sequence alignment and comparison with other BuVs, we detected 2 potential splice sites in the ORF1/ORF2 junction, a potential donor site (AG↓GT) at nt 1931 and an acceptor site (AG↓G) at nt 2115. The putative VP1 sequence started at the end of ORF1 at nt 1906, upstream of the splice donor site at nt 1931 (Figure 1, panel B). We found the phospholipase A2 (PLA2) motif (Figure 1, panel A), with its highly conserved calcium binding site (YLGPG), in the main ORF of VP1. The phospholipase catalytic residues (HD and D) were present at amino acid positions 41–42 and 63 (Figure 1, panel A). The VP1 showed <67.2% aa identity to other genera of the family Parvovirinae, including its closest relative in the Parvovirus genus (Technical Appendix[PDF - 138 KB - 1 page]). The N terminus of the CBuV VP2 protein contained a glycine-rich sequence (GGGGGGGSGVG) that was also present in other parvoviral VP2 proteins (Figure 1, panel A).
We successfully determined the complete coding genome sequence of 2 additional CBuV strains of enteric origin, HUN/2012/22 (GenBank accession no. MF198245) and HUN/2012/126 (GenBank accession no. MF198246); the sequence was 4,219 nt (4,463 and 4,308 nt with the partial UTRs, respectively). Overall, the 3 CBuV strains displayed 99.8%–99.9% nt identity to each other, with only 1 nonsilent mutation in the NS1 protein and 1 nonsilent mutation in the VP2 protein.
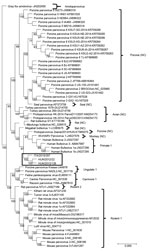
Figure 2. Capsid-based phylogenetic tree displaying the diversity of protoparvoviruses. The protoparvoviruses officially recognized by the International Committee on Taxonomy of Viruses are included, along with nonclassified (NC) protoparvoviruses. The tree was generated...
Upon phylogenetic analysis (Figure 2), the CBuV strains segregated into a well-defined group (bootstrap value 100), encompassing parvoviruses identified in rats, bats, pigs, and primates. The closest relatives within this group were parvoviruses from humans, commonly called human bufaviruses (16,17), and parvoviruses detected in monkeys (Macaca mulatta) (18) and pigs (19).
Screening in PCR of Canine Samples
Respiratory Samples
Molecular screening by qPCR detected CBuV DNA in 10/58 (17.2%) oropharyngeal swabs and 15/58 (25.8%) nasal swabs from collection RIS. In total, 31% (18/58) of the animals tested positive for CBuV in either the pharyngeal or nasal sample, whereas 12.1% (7/58) were positive in both the oral and nasal swab sample. We did not detect CBuV DNA in the NOP samples of the control group (0/90) (collection RIA). The viral loads of collection RIS ranged from 4.91 × 101 to 8.78 × 103 DNA copies/10 μL of template (mean 8.73 × 102 DNA copies; median 2.91 × 102 DNA copies). We found a statistically significant difference for CBuV prevalence in oropharyngeal (odds ratio [OR] 6, 95% CI 1.6–23) and nasal (OR 10.1, 95% CI 2.7–36.8) swab specimens when we compared animals with and without clinical signs (p<0.05 for both comparisons).
We reanalyzed the results by age-based cohorts of animals (0–6 and 7–12 months). In the 0–6-month group, only 1/17 (5.8%) animals were positive; 17/41 (41.4%) tested positive in the 7–12-month group. However, this difference was not statistically significant (p>0.05).
We also screened the NOP swabs of the litter infected by strain ITA/2011/297-15 in qPCR. We detected CBuV DNA in all 3 puppies, with viral titers as high as 2.79 × 102, 1.01 × 103 DNA, and 3.77 × 103 copies/10 μL of template.
Enteric Samples
Molecular screening by qPCR revealed CBuV DNA in 26/81 (32.1%) stools or rectal swabs from collection EIS and 15/78 (19.2%) samples of collection EIA. The viral loads of collection EIS ranged from 4.91 × 101 to 8.78 × 103 DNA copies/10 μL of template (mean 4.21 × 102 DNA copies; median 1.38 × 102 DNA copies). The viral loads of collection EIA ranged from 5.10 × 101 to 4.34 × 102 DNA copies per 10 μL of template (mean 1.76 × 102 DNA copies; median 1.48 × 102 DNA copies). We found no statistically significant difference between enteric signs and the presence of CBuV DNA in stools or rectal swabs.
We detected CBuV DNA in 8/20 (40%) enteric samples from collection EHS and 19/40 (47.5%) samples from collection EHA. The viral loads of collection EHS ranged from 1.31 × 101 to 5.42 × 103DNA copies/10 μL of template (mean 6.55 × 102 DNA copies; median 3.99 × 101 DNA copies). The viral loads of collection EHA ranged from 1.21 × 101 to 2.57 × 1010 DNA copies/10 μL of template (mean 1.35 × 109 DNA copies; median 2.92 × 102 DNA copies). We found no statistically significant difference between enteric signs and the presence of CBuV DNA in the samples from Hungary.
We subjected samples containing genome copies >103 DNA copies/10 μL of template to PCR amplification with primers CPPV 165F and CPPV 371R, which amplify a 207-nt fragment of VP2. We successfully sequenced 12 samples, yielding amplicons of the expected size in PCR and confirming the specificity of the qPCR.
Virus Cultivation
We visually inspected the inoculated monolayers of A-72, MDCK, and WRCC cells after 5 serial passages and monitored virus titer in cellular supernatant by qPCR. We did not observe viral growth in any of the cells.
We report the detection and genomic characterization of CBuV, a novel canine protoparvovirus, from a small outbreak of CIRD. In the NS1 gene, CBuV displayed low nt (24.1%–69.4%) and aa (19.3%–51.4%) identities compared with other protoparvoviruses. Current International Committee on Taxonomy of Viruses criteria for classification of parvoviruses into the same species require >85% aa identity in the NS1 protein; on this basis, CBuV could be classified as a new parvovirus species. The closest relatives to CBuV were protoparvoviruses identified in primates and other mammals (16–19), commonly termed as bufaviruses. Bufaviruses were first identified in 2012 in Burkina Faso in fecal samples from a child with enteric signs (16). Similar bufaviruses have been subsequently identified in different species of domestic and wild animals (18–20). CBuV exhibited the most similarity in NS1 (66.9%–69.4% nt and 47.2%–51.4% aa) and VP1 (66%–68.2% nt and 62.5%–67.2% aa) to bufaviruses identified in primates. In addition, the genetic organization of CBuV resembled that of primate bufaviruses, with the conservation of a potential splicing mechanism regulating VP1 translation. On the other hand, CBuV was genetically more distantly related to CPV-2, showing only 56% nt and 40.6% aa identity in the NS1 and 42.6% nt and 33.4% aa identity in the VP1 between the 2 canine protoparvoviruses. Because we were not able to adapt the virus to in vitro growth in different canine cell lines, we could not assess whether there is an antigenic relationship between the 2 canine protoparvoviruses, CBuV and CPV-2, in cross-neutralization studies.
A small case–control study on samples of enteric and respiratory origin in puppies and young dogs (<1 year of age) revealed that CBuV appeared significantly more common in NOP swab samples from dogs with acute CIRD. We detected CBuV DNA in 17.2% (10/58) of oropharyngeal swab specimens and 25.8% (15/58) of nasal swab specimens from collection RIS. In total, 31% (18/58) of the animals with CIRD tested positive for CBuV, whereas the virus was not detectable in respiratory samples of 90 animals without clinical signs. When we analyzed the results of collection RIS by age-based cohorts, we observed an increased prevalence (41.4%, 17/41) in the 7- to 12-month age group, whereas only 1/17 animals (5.8%) in the 0- to 6-month age group tested positive for CBuV. We also found a high prevalence of CBuV DNA in canine stool samples, although we observed no substantial difference between dogs with enteric disease and clinically healthy dogs. These findings indicate that CBuVs are a common component of the canine fecal virome.
With certain exceptions, it has been difficult to demonstrate a clear association of many potential pathogens with CIRD in either epidemiologic studies or experimental infections. Our study could not provide conclusive evidence for a role of this novel virus in CIRD.
CIRD or kennel cough has a multiagent etiology, with >1 agent (viruses or bacteria) involved sequentially or synergistically to cause disease (23). Pathogens commonly associated with CIRD include CAV-2, canine parainfluenza virus, and Bordetella bronchiseptica (12,24,25). Less commonly, CHV-1 can cause respiratory disease (26). CAV-1 and canine distemper virus infections are also associated with respiratory disease but are usually responsible for systemic disease (26,27).
In recent years, other emerging agents have been associated with CIRD, including canine respiratory coronavirus (27,28), canine pneumovirus (29), nonprimate canine hepacivirus (30), CBoVs (6), Mycoplasma cynos (31), and Streptococcus equi subsp. zooepidemicus (32,33). In addition, thus far, >5 strains of influenza virus have been identified in dogs: the equine-derived H3N8 virus, the human-derived H1N1 virus, and the avian-like H3N2, H3N1, and H5N2 viruses (34,35).
Of interest, the titer of CBuV in the NOP samples was not high, with a median value of 2.91 × 102 DNA copies/10 μL. This low level of virus shedding in NOP secretions was difficult to interpret, and intrinsic properties of the virus, or the dynamics of virus shedding at the time of sampling, or both could account for it. For comparison, a human parvovirus associated with respiratory disease, human bocaparvovirus 1, can be shed at titers as high as 3.9 × 1011 copies/mL in NOP samples (36). It is worth noting that we also found high loads of CBuV (<2.57 × 1010 DNA copies/10 μL of template) in 4 fecal samples that substantially exceeded the median value (2.92 × 102 DNA copies/10 μL) of collection EHA.
To date, scientists have searched for bufaviruses almost exclusively in fecal samples and have detected them in diarrheal stools of patients of all ages worldwide. The prevalence of bufaviruses in human patients ranges from 0.3% to 4%, and their etiologic role in enteric or extraenteric diseases remains uncertain (16,17,37–41). Bufaviruses have also been found in other mammalian hosts, including wild and captive nonhuman primates, swine, shrews, rats, bats, and fur seals (18–20,42–46). Of interest, bufaviruses have also been detected in the serum and spleens of monkeys and in the spleens of shrews (18,44) and in a unique NOP sample of 955 human patients with lower respiratory tract signs (47), suggesting the possibility of extraintestinal or systemic infections. In this study, we have also reconstructed the genome sequence of 2 CBuV strains detected in canine fecal samples. By genome comparison, we observed only 2 aa differences between the CBuV strains of respiratory and enteric origin, although the viruses were identified from animals of different geographic origin (i.e., Italy and Hungary). Although our findings corroborate earlier evidence that bufaviruses can target extraintestinal tissues and organs, only animal experiments or detailed observational studies can fully address this issue.
In conclusion, the advancement of techniques available for pathogen discovery is quickly broadening the list of potential canine infectious agents. Understanding in more depth the effects of those agents on canine health will be pivotal to implementing future strategies for prophylaxis, chiefly for complex diseases like CIRD.
Dr. Martella is a full professor in the Department of Veterinary Medicine, University of Bari, Italy. His research is focused on diagnosis, epidemiology, and characterization of human and animal viruses, with particular interest in zoonotic viruses.
Acknowledgments
We thank L.E. Carmichael for reading and critical revision of the manuscript.
Financial support was obtained from the grant IZS PB 2/15 RC by the Italian Ministry of Health and the Momentum Program awarded by the Hungarian Academy of Sciences. S.M. was the recipient of the Bolyai scholarship.
References
- Cotmore SF, Agbandje-McKenna M, Chiorini JA, Mukha DV, Pintel DJ, Qiu J, et al. The family Parvoviridae. Arch Virol. 2014;159:1239–47. DOIPubMed
- Binn LN, Lazar EC, Eddy GA, Kajima M. Recovery and characterization of a minute virus of canines. Infect Immun. 1970;1:503–8.PubMed
- Decaro N, Amorisco F, Lenoci D, Lovero A, Colaianni ML, Losurdo M, et al. Molecular characterization of Canine minute virus associated with neonatal mortality in a litter of Jack Russell terrier dogs. J Vet Diagn Invest. 2012;24:755–8. DOIPubMed
- Appel MJG, Scott FW, Carmichael LE. Isolation and immunisation studies of a canine parco-like virus from dogs with haemorrhagic enteritis. Vet Rec. 1979;105:156–9. DOIPubMed
- Decaro N, Buonavoglia C. Canine parvovirus—a review of epidemiological and diagnostic aspects, with emphasis on type 2c. Vet Microbiol. 2012;155:1–12. DOIPubMed
- Kapoor A, Mehta N, Dubovi EJ, Simmonds P, Govindasamy L, Medina JL, et al. Characterization of novel canine bocaviruses and their association with respiratory disease. J Gen Virol. 2012;93:341–6. DOIPubMed
- Li L, Pesavento PA, Leutenegger CM, Estrada M, Coffey LL, Naccache SN, et al. A novel bocavirus in canine liver. Virol J. 2013;10:54. DOIPubMed
- Lau SK, Woo PC, Yeung HC, Teng JL, Wu Y, Bai R, et al. Identification and characterization of bocaviruses in cats and dogs reveals a novel feline bocavirus and a novel genetic group of canine bocavirus. J Gen Virol. 2012;93:1573–82. DOIPubMed
- Parrish CR, O’Connell PH, Evermann JF, Carmichael LE. Natural variation of canine parvovirus. Science. 1985;230:1046–8. DOIPubMed
- Parrish CR, Aquadro CF, Strassheim ML, Evermann JF, Sgro JY, Mohammed HO. Rapid antigenic-type replacement and DNA sequence evolution of canine parvovirus. J Virol. 1991;65:6544–52.PubMed
- Buonavoglia C, Martella V, Pratelli A, Tempesta M, Cavalli A, Buonavoglia D, et al. Evidence for evolution of canine parvovirus type 2 in Italy. J Gen Virol. 2001;82:3021–5. DOIPubMed
- Decaro N, Mari V, Larocca V, Losurdo M, Lanave G, Lucente MS, et al. Molecular surveillance of traditional and emerging pathogens associated with canine infectious respiratory disease. Vet Microbiol. 2016;192:21–5. DOIPubMed
- Mihalov-Kovács E, Gellért Á, Marton S, Farkas SL, Fehér E, Oldal M, et al. Candidate new rotavirus species in sheltered dogs, Hungary. Emerg Infect Dis. 2015;21:660–3. DOIPubMed
- Scotto-Lavino E, Du G, Frohman MA. 5′ end cDNA amplification using classic RACE. Nat Protoc. 2006;1:2555–62. DOIPubMed
- Decaro N, Amorisco F, Desario C, Lorusso E, Camero M, Bellacicco AL, et al. Development and validation of a real-time PCR assay for specific and sensitive detection of canid herpesvirus 1. J Virol Methods. 2010;169:176–80. DOIPubMed
- Phan TG, Vo NP, Bonkoungou IJ, Kapoor A, Barro N, O’Ryan M, et al. Acute diarrhea in West African children: diverse enteric viruses and a novel parvovirus genus. J Virol. 2012;86:11024–30. DOIPubMed
- Yahiro T, Wangchuk S, Tshering K, Bandhari P, Zangmo S, Dorji T, et al. Novel human bufavirus genotype 3 in children with severe diarrhea, Bhutan. Emerg Infect Dis. 2014;20:1037–9. DOIPubMed
- Handley SA, Thackray LB, Zhao G, Presti R, Miller AD, Droit L, et al. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012;151:253–66. DOIPubMed
- Hargitai R, Pankovics P, Kertész AM, Bíró H, Boros Á, Phan TG, et al. Detection and genetic characterization of a novel parvovirus distantly related to human bufavirus in domestic pigs. Arch Virol. 2016;161:1033–7. DOIPubMed
- Sasaki M, Gonzalez G, Wada Y, Setiyono A, Handharyani E, Rahmadani I, et al. Divergent bufavirus harboured in megabats represents a new lineage of parvoviruses. Sci Rep. 2016;6:24257. DOIPubMed
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–51.PubMed
- Phan TG, Sdiri-Loulizi K, Aouni M, Ambert-Balay K, Pothier P, Deng X, et al. New parvovirus in child with unexplained diarrhea, Tunisia. Emerg Infect Dis. 2014;20:1911–3. DOIPubMed
- Buonavoglia C, Martella V. Canine respiratory viruses. Vet Res. 2007;38:355–73. DOIPubMed
- Joffe DJ, Lelewski R, Weese JS, Mcgill-Worsley J, Shankel C, Mendonca S, et al. Factors associated with development of Canine Infectious Respiratory Disease Complex (CIRDC) in dogs in 5 Canadian small animal clinics. Can Vet J. 2016;57:46–51.PubMed
- Schulz BS, Kurz S, Weber K, Balzer HJ, Hartmann K. Detection of respiratory viruses and Bordetella bronchiseptica in dogs with acute respiratory tract infections. Vet J. 2014;201:365–9. DOIPubMed
- Decaro N, Martella V, Buonavoglia C. Canine adenoviruses and herpesvirus. [viii.]. Vet Clin North Am Small Anim Pract. 2008;38:799–814, viii. DOIPubMed
- Decaro N, Desario C, Elia G, Mari V, Lucente MS, Cordioli P, et al. Serological and molecular evidence that canine respiratory coronavirus is circulating in Italy. Vet Microbiol. 2007;121:225–30. DOIPubMed
- Erles K, Toomey C, Brooks HW, Brownlie J. Detection of a group 2 coronavirus in dogs with canine infectious respiratory disease. Virology. 2003;310:216–23. DOIPubMed
- Renshaw RW, Zylich NC, Laverack MA, Glaser AL, Dubovi EJ. Pneumovirus in dogs with acute respiratory disease. Emerg Infect Dis. 2010;16:993–5. DOIPubMed
- Kapoor A, Simmonds P, Gerold G, Qaisar N, Jain K, Henriquez JA, et al. Characterization of a canine homolog of hepatitis C virus. Proc Natl Acad Sci U S A. 2011;108:11608–13. DOIPubMed
- Chalker VJ, Owen WM, Paterson C, Barker E, Brooks H, Rycroft AN, et al. Mycoplasmas associated with canine infectious respiratory disease. Microbiology. 2004;150:3491–7. DOIPubMed
- Chalker VJ, Brooks HW, Brownlie J. The association of Streptococcus equi subsp. zooepidemicus with canine infectious respiratory disease. Vet Microbiol. 2003;95:149–56. DOIPubMed
- Priestnall SL, Erles K, Brooks HW, Cardwell JM, Waller AS, Paillot R, et al. Characterization of pneumonia due to Streptococcus equi subsp. zooepidemicus in dogs. Clin Vaccine Immunol. 2010;17:1790–6. DOIPubMed
- Crawford PC, Dubovi EJ, Castleman WL, Stephenson I, Gibbs EP, Chen L, et al. Transmission of equine influenza virus to dogs. Science. 2005;310:482–5. DOIPubMed
- Zhu H, Hughes J, Murcia PR. Origins and evolutionary dynamics of H3N2 canine influenza virus. J Virol. 2015;89:5406–18. DOIPubMed
- Zhou L, Zheng S, Xiao Q, Ren L, Xie X, Luo J, et al. Single detection of human bocavirus 1 with a high viral load in severe respiratory tract infections in previously healthy children. BMC Infect Dis. 2014;14:424. DOIPubMed
- Väisänen E, Kuisma I, Phan TG, Delwart E, Lappalainen M, Tarkka E, et al. Bufavirus in feces of patients with gastroenteritis, Finland. Emerg Infect Dis. 2014;20:1077–9. DOIPubMed
- Smits SL, Schapendonk CM, van Beek J, Vennema H, Schürch AC, Schipper D, et al. New viruses in idiopathic human diarrhea cases, the Netherlands. Emerg Infect Dis. 2014;20:1218–22. DOIPubMed
- Chieochansin T, Vutithanachot V, Theamboonlers A, Poovorawan Y. Bufavirus in fecal specimens of patients with and without diarrhea in Thailand. Arch Virol. 2015;160:1781–4. DOIPubMed
- Altay A, Yahiro T, Bozdayi G, Matsumoto T, Sahin F, Ozkan S, et al. Bufavirus genotype 3 in Turkish children with severe diarrhoea. Clin Microbiol Infect. 2015;21:965.e1–4. DOIPubMed
- Huang DD, Wang W, Lu QB, Zhao J, Guo CT, Wang HY, et al. Identification of bufavirus-1 and bufavirus-3 in feces of patients with acute diarrhea, China. Sci Rep. 2015;5:13272. DOIPubMed
- Yang S, Liu D, Wang Y, Qu F, He Y, Sun Z, et al. Bufavirus Protoparvovirus in feces of wild rats in China. Virus Genes. 2016;52:130–3. DOIPubMed
- Kluge M, Campos FS, Tavares M, de Amorim DB, Valdez FP, Giongo A, et al. Metagenomic survey of viral diversity obtained from feces of subantarctic and South American fur seals. PLoS One. 2016;11:e0151921. DOIPubMed
- Sasaki M, Orba Y, Anindita PD, Ishii A, Ueno K, Hang’ombe BM, et al. Distinct lineages of bufavirus in wild shrews and nonhuman primates. Emerg Infect Dis. 2015;21:1230–3. DOIPubMed
- Liu L, Schwarz L, Ullman K, Ahola H, Qiu Y, Ma Z, et al. Identification of a novel bufavirus in domestic pigs by a viral metagenomic approach. J Gen Virol. 2016;97:1592–6. DOIPubMed
- Kemenesi G, Dallos B, Görföl T, Estók P, Boldogh S, Kurucz K, et al. Genetic diversity and recombination within bufaviruses: Detection of a novel strain in Hungarian bats. Infect Genet Evol. 2015;33:288–92. DOIPubMed
- Väisänen E, Paloniemi M, Kuisma I, Lithovius V, Kumar A, Franssila R, et al. Epidemiology of two human protoparvoviruses, bufavirus and tusavirus. Sci Rep. 2016;6:39267. DOIPubMed
Figures
Table
Technical Appendix
Cite This ArticleOriginal Publication Date: 5/3/2018






















.png)











No hay comentarios:
Publicar un comentario